Information on the MALDI Quick Microbiological Identification Test
The MALDI-TOF MS-based test for identifying microbes is totally different from the conventional identification test.
What is the microbial identification test by MALDI-TOF MS?
In MALDI-TOF MS, the sample is ionized in a process called matrix-assisted laser desorption/ionization (MALDI) and identified by time-of-flight mass spectrometry (TOF MS).
To identify microbial samples (or isolates), the mass spectrum from the sample is obtained by MALDI-TOF MS and compared with the mass spectra of microbes in the database.
As this method is rapid and produces results highly similar to the sequencing results of the 16S rRNA gene, many diagnostic laboratories have started to identify isolates with MALDI-TOF MS.
At the ICLAS Monitoring Center, identification is made against the MALDI Biotyper (Bruker Daltonics K.K.) database, and a quality controlled database of bacteria from animals that we independently established.
MALDI Biotyper Libraries
MBT 8468 MSP Library (2019 on release)
- Gram negative bacteria 1,266 species
- Gram positive bacteria 1,450 species
- Yeast 210 species
- Filamentous fungi 43 species
MBT Filamentous Fungi Library 3.0 (2019 on release)
- Filamentous fungi 180 species / species groups
Samples
Specimens: Bacteria, yeasts and filamentous fungi
Preparation: Fresh isolates (only bacteria and yeasts) that have been cultured to a visible colony size on the plate medium. It has to be a pure culture isolates.
Filamentous fungi do not need to be fresh isolates.
Report format
The sample's protein spectrum is screened against the database of spectra from known microbes to match the protein peak pattern for identification.
Accordingly, if protein peak patterns are similar between the sample and a microbe in the database, that particular microbe will be identified in the MALDI Biotyper Classification Results even if it is not the actual microbe in the sample.
Figure: Example of report form
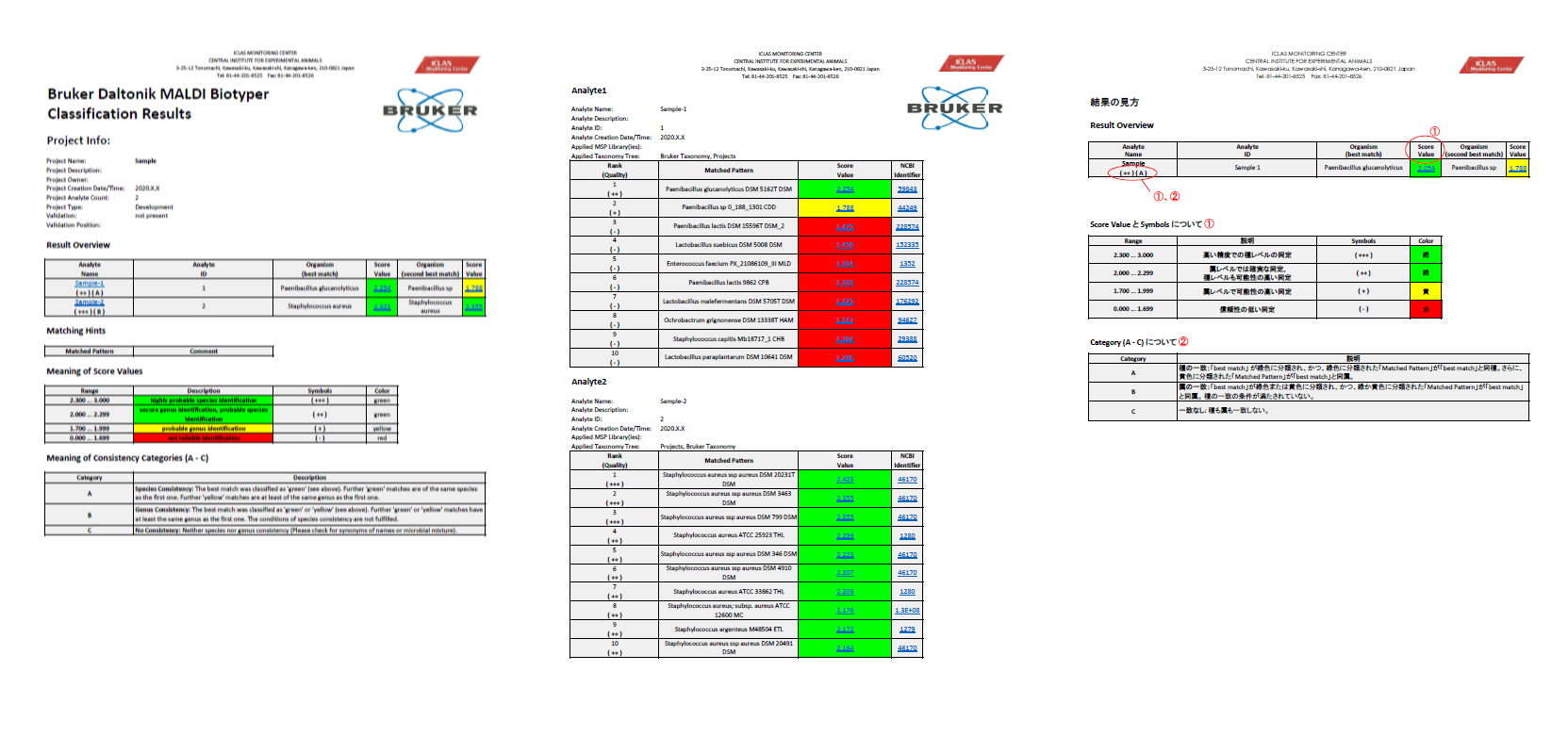 Standard MALDI Biotyper report format Notion of results is here
Standard MALDI Biotyper report format Notion of results is here
